Teaching in courses:
Protein Sorting

Henrik Nielsen, Associate Professor
Different compartments in eukaryotic cells contain different sets of proteins. Even prokaryotic cells have differences between cytosolic proteins, membrane proteins, and secretory proteins. So how does the cell know where to put which proteins? In other words: Where is the information that makes protein sorting possible? Proteins have intrinsic signals that govern their transport and localization in the cell — this was the discovery that earned Günter Blobel the Nobel prize in Physiology or Medicine 1999. In the protein sorting group, we aim to characterize and predict these intrinsic signals — the “zip codes” of proteins.
We use modern machine learning methods, including deep learning, to find and predict the “zip codes” from amino acid sequences. We have been using artificial neural networks since the 1990s, but during the latest decade, neural networks have experienced a revival since deep learning became practically possible with the implementations of Convolutional Neural Networks and Long Short-Term Memory. More recently, the field has borrowed inspiration from the Natural Language Processing field, and several of our newer tools use protein language models.
There is a huge interest in protein sorting prediction from both the academic and industrial environments. E.g., the biotechnological industry is interested in protein secretion for production of recombinant proteins, while the pharmaceutical industry is interested in cell surface proteins as drug targets.
Projects
SignalP
Signal Peptides and their cleavage sites in all domains of life
The most well-known and ubiquitous protein “zip code” is the secretory signal peptide, which is found in both eukaryotes, bacteria, archaea, and viruses. In prokaryotes, this is a signal for export across the cell membrane, while in eukaryotes, it signals export across the endoplasmic reticulum (ER) membrane. The protein sorting group is responsible for the SignalP method for predicting signal peptides and their cleavage sites. The SignalP web server, originally launched in 1996, is used more than 1,000 times daily, and thousands of users have downloaded the program for use at their own computers. SignalP version 6.0 from 2022 is based on protein language models and predicts all five types of signal peptides in prokaryotes. The articles about SignalP have been cited more than 20,000 times in total; see Henrik Nielsen's Google Scholar, Scopus, and Web of Science pages.
TargetP
Transit Peptides for protein import into chloroplasts and mitochondria
Transit peptides are “zip codes” that signal import into mitochondria or plastids (e.g. chloroplasts). The TargetP web server predicts N-terminal sorting signals including transit peptides and signal peptides, and classifies eukaryotic proteins as belonging to mitochondria, plastids, secretory pathway, or other destinations. TargetP version 2.0 from 2019 is based on deep learning and includes prediction of thylakoid luminal transit peptides in chloroplasts.
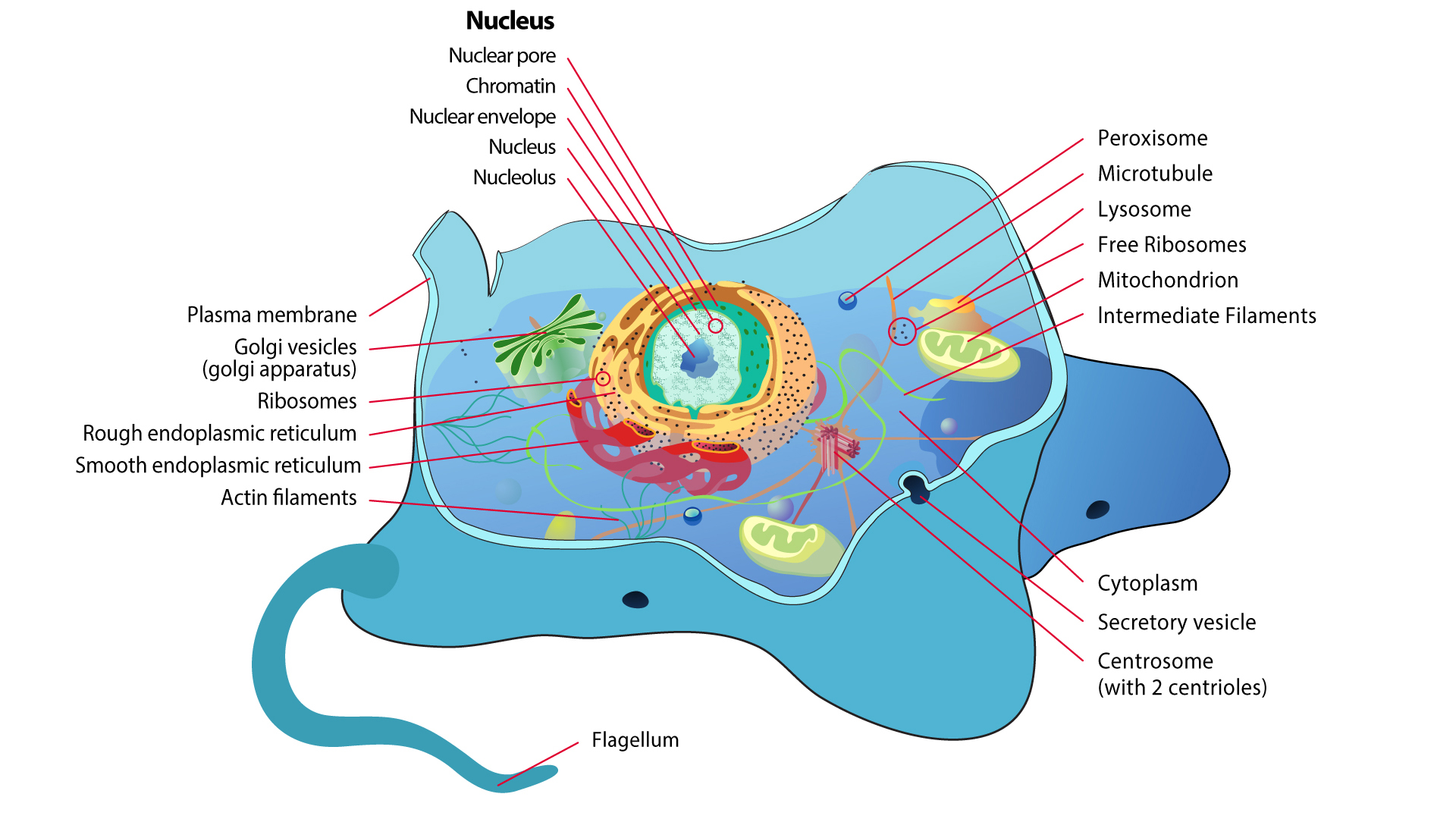
DeepLoc
Eukaryotic protein subcellular localization in 10 categories
The DeepLoc web server classifies eukaryotic proteins as belonging to Nucleus, Cytoplasm, Extracellular, Mitochondrion, Cell membrane, Endoplasmic reticulum, Chloroplast, Golgi apparatus, Lysosome/Vacuole or Peroxisome. Version 2.0 from 2022 is based on protein language models and is able to predict dual locations and some sorting signals.
NetSurfP
Protein secondary structure and solvent accessibility
The NetSurfP web server predicts one-dimensional aspects of protein structure, namely the surface accessibility, secondary structure, disorder, and φ/ψ dihedral angles of each amino acid residue. Version 3.0 from 2022 is based on protein language models and runs much faster than version 2.0 since it does not depend on building sequence profiles from a database.
NetGPI
GPI-anchoring in eukaryotic proteins
GPI-anchoring is a post-translational modification responsible for attaching many eukaryotic proteins to the outer face of the plasma membrane. A C-terminal “zip code” is recognized, cleaved, and replaced by a glycosylphosphatidylinositol group that anchors the protein to the lipid membrane. The NetGPI web server predicts the presence and ω-sites (where cleavage and modification happens) of GPI-anchor signals.
NetSolP
Solubility and usability of proteins expressed in E. coli
When proteins are expressed in a production host such as Escherichia coli, the success of expression and the solubility of the protein product depend on many factors, one of them being the sequence of the protein. The NetSolP web server predicts the solubility and usability for purification of proteins expressed in E. coli from the amino acid sequence.
NetStart
Eukaryotic start codons in nucleotide sequences
In a eukaryotic messenger RNA, the start codon (where translation begins) is not necessarily the first occurrence of AUG. The NetStart web server predicts which AUG triplet in an mRNA sequence is the start codon.
Henrik Nielsen Associate Professor Department of Health Technology Phone: +45 45252098 henni@dtu.dk