Teaching in courses:
Research Group
Isoform Analysis Group

Group leader: Kristoffer Vitting-Seerup
The Isoform Analysis Group aims to inspire and enable gene isoform analysis across all the health and natural sciences. We seek to do this by developing the bioinformatic infrastructure necessary to analyze isoforms (enable) and use it to show the importance of isoforms in human health and disease (inspire).
Background
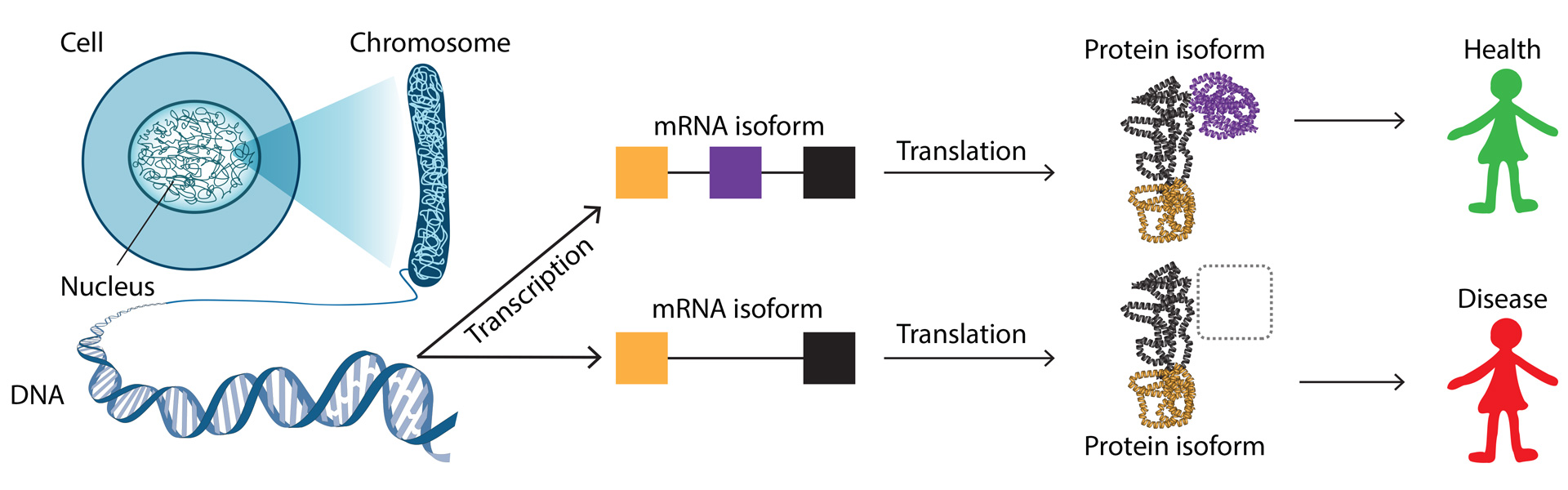
Alternative splicing, alternative transcription start sites, and alternative termination sites collectively augment the functional repertoire of genomes, facilitating the generation of diverse isoforms from a single gene (illustrated in Figure 1). The choice of isoform holds tremendous biological relevance; for instance, pivotal genes in apoptosis have isoforms—one promoting and the other inhibiting apoptosis. In other words, only measuring gene-level changes is rather useless for these genes – we need to know about the isoforms.
We now know of hundreds of such lab-validated cases where individual isoforms drive biological changes. It is, therefore, astonishing that only ~10% of researchers across a wide range of bioinformatics fields seem to be analyzing isoforms (Figure 2), indicating we are currently neither seeing nor understanding the full picture.
The Challenge
If we generalize from Figure 2 to other research fields and diseases, it becomes abundantly clear that no single group or consortium could ever do this work alone! There is simply too much work to be done in too many research fields.
Therefore, the only viable solution is to inspire and enable other researchers to analyze isoforms.
Inspiring isoform analysis
To inspire isoform analysis, we have so far taken a bioinformatic approach and reanalyzed hundreds of high-throughput datasets covering many different high-throughput approaches. For each dataset and data modality, we investigate the additional biological information hidden at the isoform level. This work, which we are in the process of publishing, shows that isoforms are a central part of the cellular response to any intervention (experimental, disease, or otherwise) independent of which research field interrogated. Summarizing across these studies we find that ~50% of the cellular response is, at least to some extent, mediated by the isoform level changes that are currently overlooked. With this work, we hope to inspire researchers worldwide to analyze isoforms within their scope of interest.
Enabling isoform analysis
While our work highlights the broad impact of isoform-level changes, understanding their specific biological functions remains challenging. For while we know a lot about the biological function of most genes, we know next to nothing about the biological function of the underlying isoforms. Committed to addressing this gap, our group endeavors to develop accessible methods for interpreting isoform data, empowering researchers to unlock the full potential of isoform analysis in elucidating complex biological processes.
Technological and computational approach
The Isoform Analysis Group leverages high-throughput technologies to investigate individual genes/isoforms and global profiling of cellular programs, pathways, and their dynamic changes. Our core focus includes both bulk/single-cell RNA sequencing and bulk/single-cell proteomics, but we utilize most high-throughput technologies when advantageous.
On the computational side, we simply use whichever tool is better suited for the task. This also includes developing our own tools, statistical models, and machine learning frameworks (including deep neural networks and generative modeling) whenever no existing tool can satisfactorily solve our problems.
Long-term vision
We believe the scientific field needs to change how it thinks about molecular and cellular biology. Although it will require a colossal effort, we must progress from the current ‘gene-centric’ research paradigm to a more nuanced ‘isoform-centric’ paradigm. The Isoform Analysis Group will work hard to make this reality possible.

Group defining papers
Mikkelsen et al. Beyond the Gene in Genetics: How Isoform-Resolved Analysis Empowers the Study of Both Common and Rare Genetic Variation.
https://doi.org/10.1101/2025.03.31.25324931- Dam et al. Expression and Splicing Mediate Distinct Biological Signals
doi.org/10.1101/2022.08.29.505720 - Vitting-Seerup et al. IsoformSwitchAnalyzeR: analysis of changes in genome-wide patterns of alternative splicing and its functional consequences
doi.org/10.1093/bioinformatics/btz247 - Vitting-Seerup et al. The Landscape of Isoform Switches in Human Cancers
doi.org/10.1158/1541-7786.mcr-16-0459
Group Leader
Kristoffer Vitting-Seerup Associate Professor Department of Health Technology krivi@dtu.dk
Dimitrios S. Kanakoglou PhD Student Department of Health Technology dimkan@dtu.dk
Chiao-Yu Hsieh PhD Student Department of Health Technology chihs@dtu.dk
Elena Iriondo Delgado Research Assistant Department of Health Technology eirde@dtu.dk
Isabella Østerlund Postdoc Department of Health Technology nisos@dtu.dk
Johanne Badsberg Overgaard PhD Student Department of Health Technology jobao@dtu.dk
Mikkel Niklas Rasmussen PhD Student Department of Applied Mathematics and Computer Science minra@dtu.dk
Zi Tian Zhen PhD Student Department of Health Technology ztizh@dtu.dk
Alumni:
Mo Zhou (Post-doc)
Chunxu Han
Project topics
In the isoform analysis group, we love working with students doing special courses and/or their BSc/MSc thesis. These projects are all about applying or developing bioinformatics/machine learning (including deep learning) methods for analyzing (single cell/bulk) RNA-seq, proteomics, or other high-throughput data. I know this is a very generic project description, but that is simply because I believe in selecting and tailoring projects to you and your interests. Examples could be “Enabling gene-set enrichment analysis of isoforms” or “Machine learning to enable an analysis of isoforms”.
If you think this sounds interesting, contact Kristoffer and:
- Describe your research interests and/or skills/topics you would like to learn
- Describe your experience with programming (R/Python)
- Mention the type of project (BSc, MSc, or special course), including number of ECTS
- Attach your grade transcripts, so we, prior to meeting with you, can select and adapt projects to your skill profile
Supervision Ideology
One of my core ambitions is to help you develop into self-sustained learners. Towards this goal, I subscribe to building confidence and independence through the “guided mastery” principle. This principle states that confidence and independence can be built through continuously succeeding on tasks of increasing complexity. In practice, this means I will be helping you a lot at the start of a project, but ultimately, my role will transition from being a primary source of knowledge to a guide with whom you can discuss ideas, interpretations, and broader concepts. Importantly, I am very adaptive in my supervision style, so please let me know how I can motivate you.