Teaching in courses:
Research Group
Cancer Systems Biology
Group leader: Elena Papaleo
Our group (Cancer System Biology, CSB) applies a system-biology view to the analyses of different sources of cancer data (e.g., gene expression, genomics, proteomics) to identify biomarkers. In addition, we complement our studies with structure-based approaches to classify cancer variants or to study interactions between cancer proteins and small molecules. Our research is applied both Pan-cancer than to specific cancer types, with the main focus on childhood and breast cancers. The group activities have been developed naturally in a translational direction, with the basic research on cancer mechanisms being the base for the more applicative and translational one.
Research Projects

Variant effects in rare diseases in the Nordic Countries
Our group is one of the partners of the ELEGANT-North Consortium (EN; Exploring Leukemia: Education Genetics and Technology; New Option for Rare Diseases Towards Health) founded by EU-Interreg, an EU-founded cooperative effort between academia and clinicians in different Nordic Countries.
In this project, we aim to study genomic alterations found in children with cancer, focusing mainly on leukemia cancer types, as a prototype of a rare disease. Rare diseases are life-threatening or severely debilitating and occur in fewer individuals, most of which are caused by known genetic alterations. The challenge, especially in the case of cancer, is that two patients with the same rare disease may have different course of the disease due to other variants in their genome that are poorly investigated.
ELEGANT NORTH provides access to transnational genomic and clinical data in the Nordic Countries to increase the number of cases that can be analyzed for a specific rare disease that can be used for the research. In particular, we are interested in applying and further developing a computational framework for variant interpretation developed in our group, i.e. MAVISp, to understand the mechanisms of action of variants found in the cohorts of patients provided by ELEGANT NORTH.
We will re-analyze pre-collected and curated data from whole genome sequencing or targeted sequencing from the hospital cohorts to prioritize poorly investigated genes and their mutations for structural studies. The framework includes a variety of computational methods based on molecular modelling and simulations to associate the corresponding functional effects to protein mutations and support the identification of the suitable experimental design for validation, for example, for proteins involved in cell death, proliferation or DNA repair.
Additionally, the analyses with the framework will generate a group of features that can be combined with the clinical data on the patient history and therapy response using machine learning approaches, which can shed light on the different patients’ outcomes, along with identifying new targetable mutations or mutations that are associated with predisposition to childhood cancer. Additionally, the most interesting mutations identified by the screening in silico will be validated experimentally using cellular models for acute lymphoblastic or myeloid leukemia with cellular assays to study the effects of the mutations on protein levels, interactions and biological readouts correlated to cancer progression (such as changes in cell proliferation, migration, DNA repair, cell death).
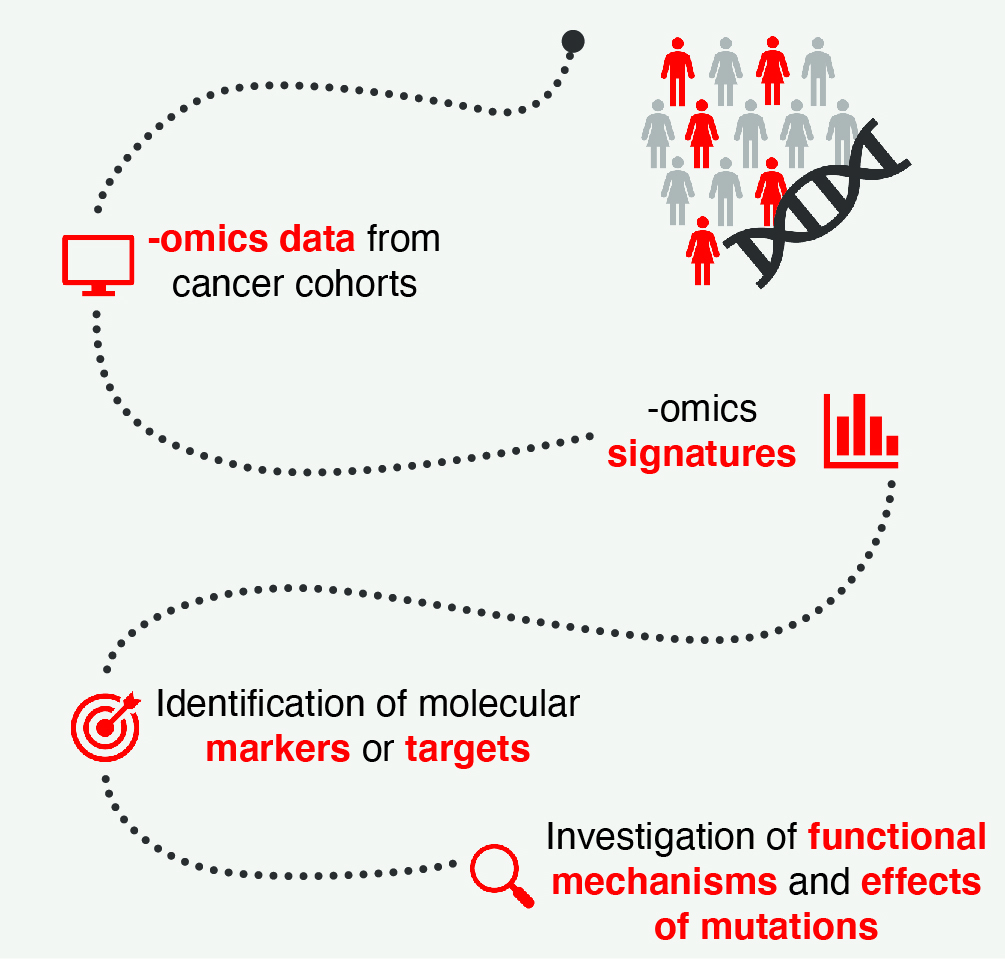
Prediction of cancer drivers and biomarkers
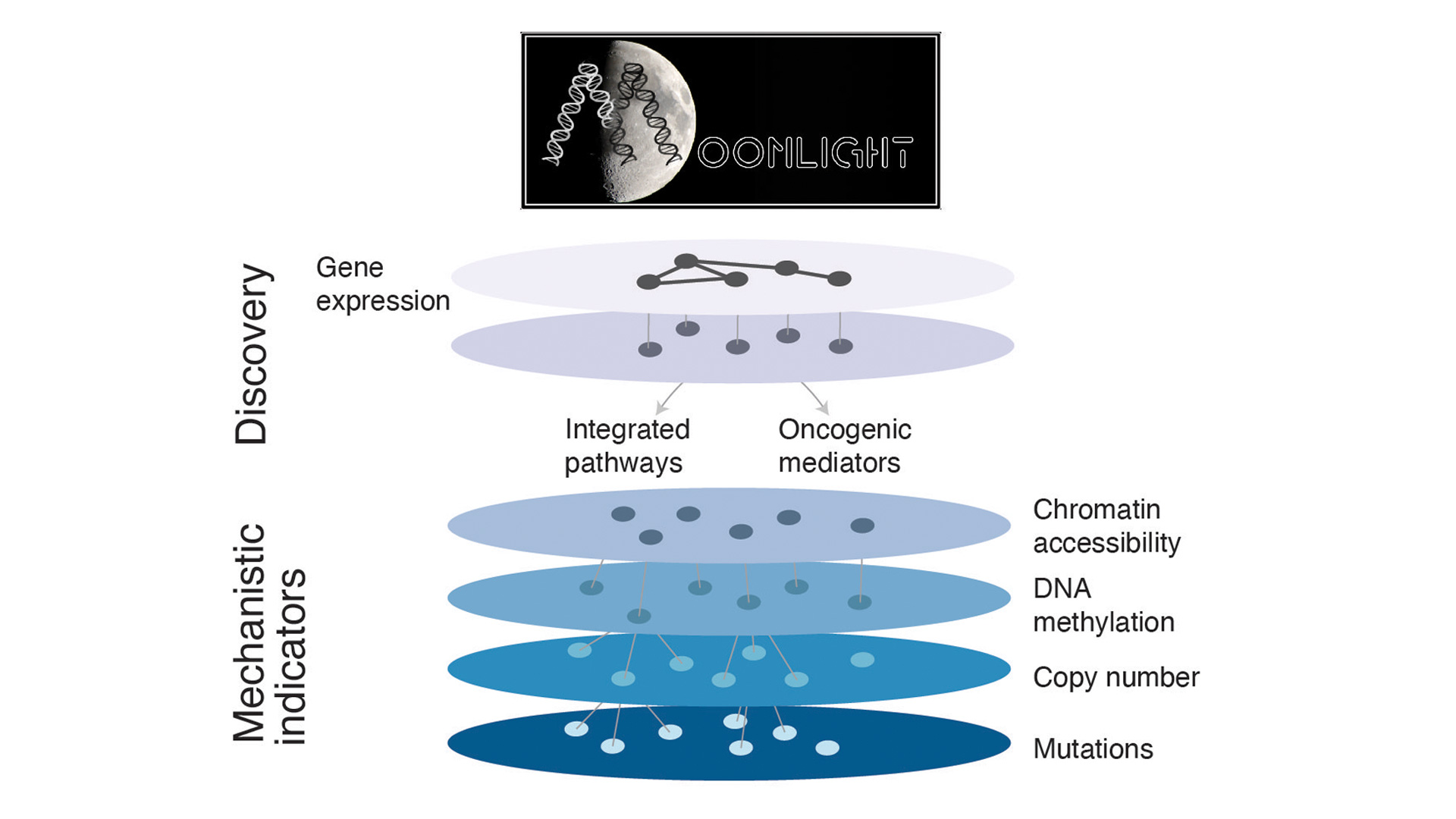
We have been developing Moonlight, a framework based on the integration of different biological data to identify driver genes in cancer and classify them in tumor suppressors and oncogenes. The framework is also used to identify dual role genes, i.e., those genes that can act both as tumor suppressors or oncogenes depending on the cellular context.
The research that we carry out with Moonlight is accompanied by other approaches for clustering and stratification of the samples depending on their gene expression, genomics, or proteomics signatures. As an example, we have been developing CAMPP (CAncer bioMarker Prediction Pipeline, for the analyses of different sources of cancer data. CAMPP includes data-management tools for missing value imputation, quality checks, normalization protocols, data clustering, differential expression, survival, and network-based analyses.
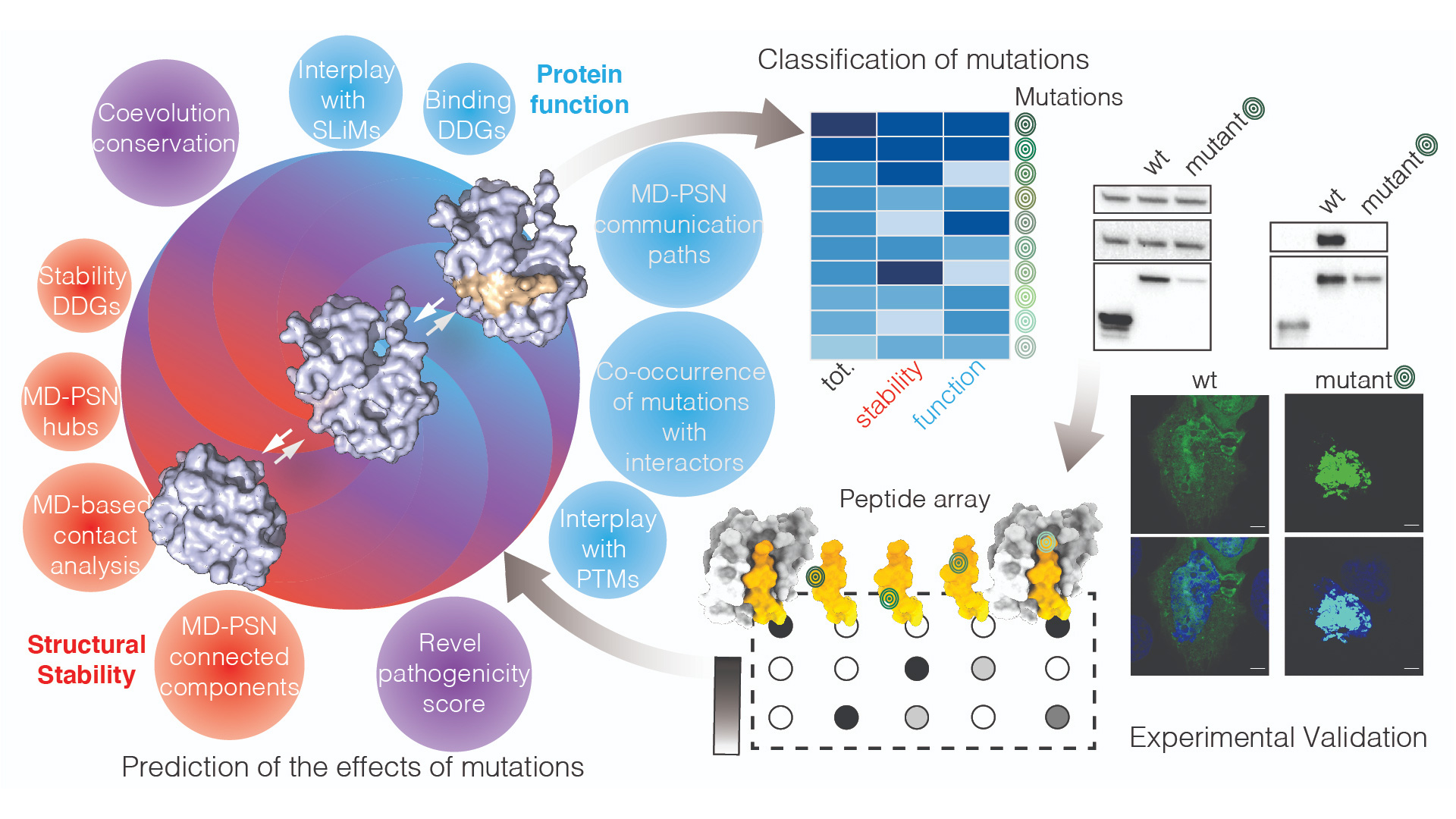
Classification and prioritization of cancer variants
We aim to provide an atlas of cancer alterations affecting the protein products that can also be consulted to prioritize variants for experimental or functional studies.
We aim to go beyond the available tools to predict pathogenic mutations based on sequence or simple structural features. These methods do not provide enough details about the mechanisms related to the alterations and how we can counteract them.
In our workflow, we aim to comprehensively account for different aspects that mutational events alter in terms of protein structure, dynamics, and function. For examples, the variants are ranked and classified according to the alteration at the level of stability of the protein product, changes in post-translational regulations, interactome, also including allosteric changes that are the results of the alteration at a distal site with respect to the functional one. As an example, we applied the workflow to the study of a key autopaghy protein and its cancer-related mutations.
Group Leader
Elena Papaleo Associate Professor Department of Health Technology elpap@dtu.dk
Collaboration
Parts of the activities are carried out in close collaboration with clinicians in Denmark and Sweden thanks to our participation in the i-COPE Consortium.
We are also affiliated with the Danish Cancer Society Research Center with the Cancer Structural Biology Laboratory (CSB), where our colleagues focus on structural biology and experiments with biochemical and cellular assays. The CSB and CBL groups work closely together with the results generated by CSB on interesting cancer targets analyzed under the lens of structural biology at CBL. CBL can also support experimental validations with biochemical assays in vitro or cellular models.
We also work closely with the University of Miami (Prof. Steven Chen and Dr. Antonio Colaprico) to develop and maintain software for the analyses of large cancer datasets.

