DTU Health Tech researchers develop a method for detection of SARS-CoV-2 RNA with the ability to be designed to detect other diseases.
The current SARS-CoV-2 RNA detection methods recommended by the World Health Organization profoundly rely on the roles of biological enzymes. High cost, stringent transportation and storage conditions as well as the global supply shortages of enzymes, limit large-scaled testing. This means that most countries have to prioritize testing on vulnerable cases, which create delay in diagnostics and identification of positive cases, which again can hamper pandemic mitigation and suppression.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) is still the gold standard for whole genome detection and has been playing a key role in controlling the Covid-19 pandemic. However, the sample-to-result takes several hours and the method necessitates a complex thermocycler instrument to raise and lower the temperature of the reaction in discrete steps.
Simpler and less expensive
Non-enzymatic isothermal amplification methods, being simpler and faster, have shown promising potentials to substitute the qRT-PCR. Although these methods perform very well when the target gene is short, they are yet to function efficiently for detection of whole genomes (long DNA or RNA targets).
"Being directly involved in improving people’s health is the ultimate dream of a biomedical researcher and we believe that the NISDA assay has given us this wonderful chance to attain that ambition"
Postdoc Mohsen Mohammadniaei
During the COVID-19 pandemic, the euro area alone experienced a 3.8% drop in GDP within the first quarter of 2020 (Eurostat 2020). Thus, developing a lower-cost methodology for pathogen detection would be highly beneficial for both patients and the healthcare systems aiming to battle future pandemics.
Associate Professor Yi Sun and Postdoc researcher Mohsen Mohammadniaei at DTU Health Tech have invented a one-pot assay, which they have named NISDA (Non-enzymatic isothermal strand displacement and amplification assay). The assay is for rapid detection of SARS-CoV-2 RNA without the need for the RNA reverse transcription step of the qRT-PCR methodology. Being one-pot, enables a single step detection routine, i.e. the user only needs to add the sample into a single tube, place it in the instrument and wait for 30 minutes to obtain the result.
The assay works at constant temperature, requires no enzymes and is based on the toehold-mediated strand displacement (TMSD) approach. TMSD is an enzyme-free molecular tool from which one strand of DNA or RNA (output) is displaced by another strand (input) to form a more stable duplex structure.
High accuracy and sensitivity
The NISDA assay was able to detect a very low concentration of RNA (10 copies/µL) in only 30 minutes. In collaboration with Hvidovre Hospital and Bispebjerg Hospital, the research team could clinically validate the NISDA assay, representing 100 % specificity as well as 96.77% and 100% sensitivity when setting up in the laboratory and hospital, respectively.
Associate Professor Yi Sun elaborates, “We exploited the TMSD approach and designed three DNA probes. One probe exchanged the whole genome to a short DNA strand and the other two probes utilized the exchanged short DNA for triggering a fluorescence signal amplification cascade reaction. The beauty of NISDA assay is its simplicity. We removed the usage of enzymes to reduce the assay cost and enhance its robustness at room temperature.”
In the assay workflow, the extracted RNA from throat swab samples is added to the reaction mixture and incubated at 42 °C for 30 minutes. The next step is fluorescence measurement, and samples with significantly higher fluorescence signals than that of the control samples (negative) are considered positive.
Towards a multiple disease diagnostics tool
“Being directly involved in improving people’s health is the ultimate dream of a biomedical researcher and we believe that the NISDA assay has given us this wonderful chance to attain that ambition”, Postdoctoral Researcher Mohsen Mohammadniaei says.
“The next step is to further design the NISDA assay for detecting different pathogens and develop a point-of-care diagnostic device for multiple disease diagnostics. Another advantage of NISDA assay is its ability to be designed for short RNA targets such as cancer biomarker microRNA. We are currently exploring different schemes for the commercialization of the NISDA assay and we are certain that the NISDA assay will become widely-known in the near future”, Associate Professor Yi Sun finishes.
Read the full paper in Nature Communication here: https://www.nature.com/articles/s41467-021-25387-9
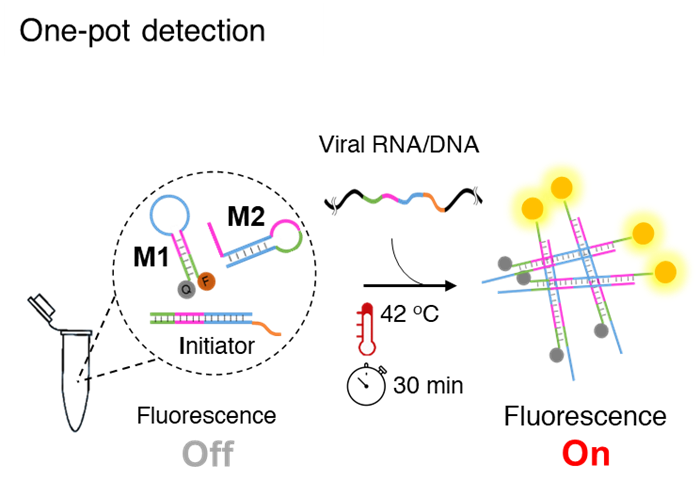
Figure:The NISDA assay comprises a single tube containing three DNA probes. After the addition of the extracted RNA from swab samples, and incubation at 42 OC for 30 min, the positive samples show higher fluorescent signals than that of the negative samples.