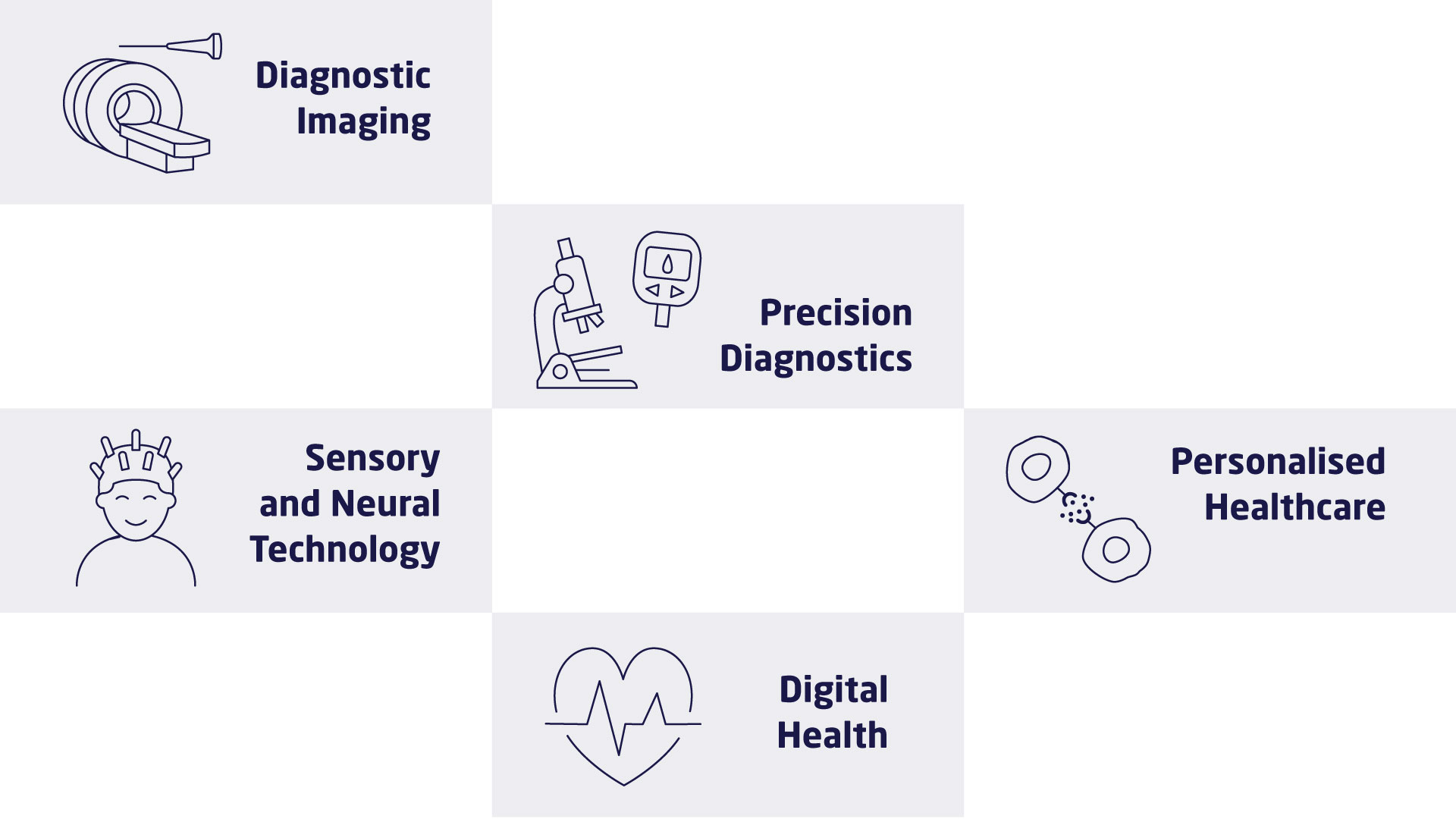
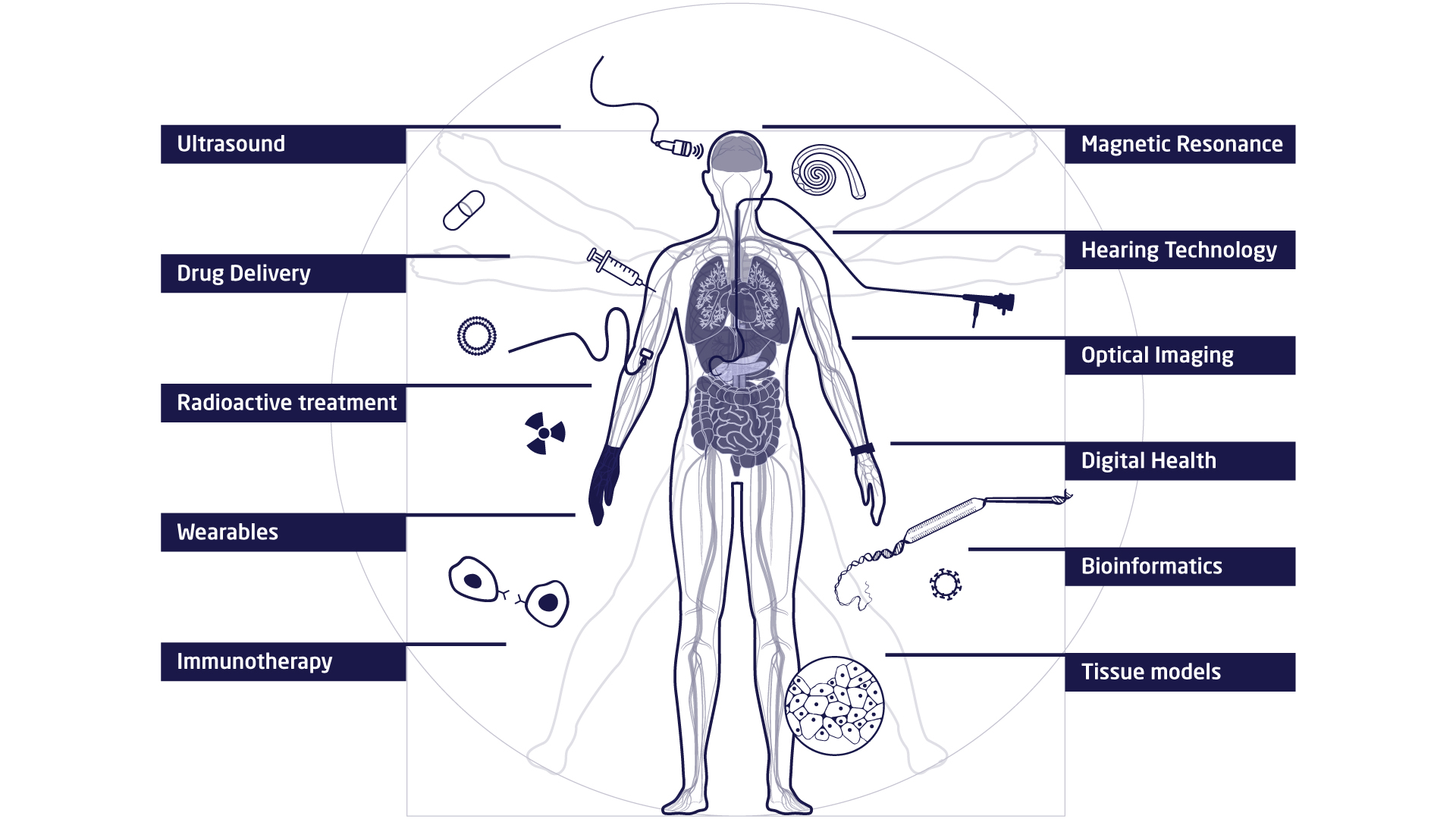
The research at DTU Health Tech is organised in research sections that each contains a number of research groups. The research sections have expertise within and across Biopharma (wet), Imaging and Diagnostics (dry) and Digital Health and Biological Modelling (data) constituting a truly interdisciplinary department. DTU Health Tech hosts a number of major research centres that include partners from industry, the health sector and academia.
Research