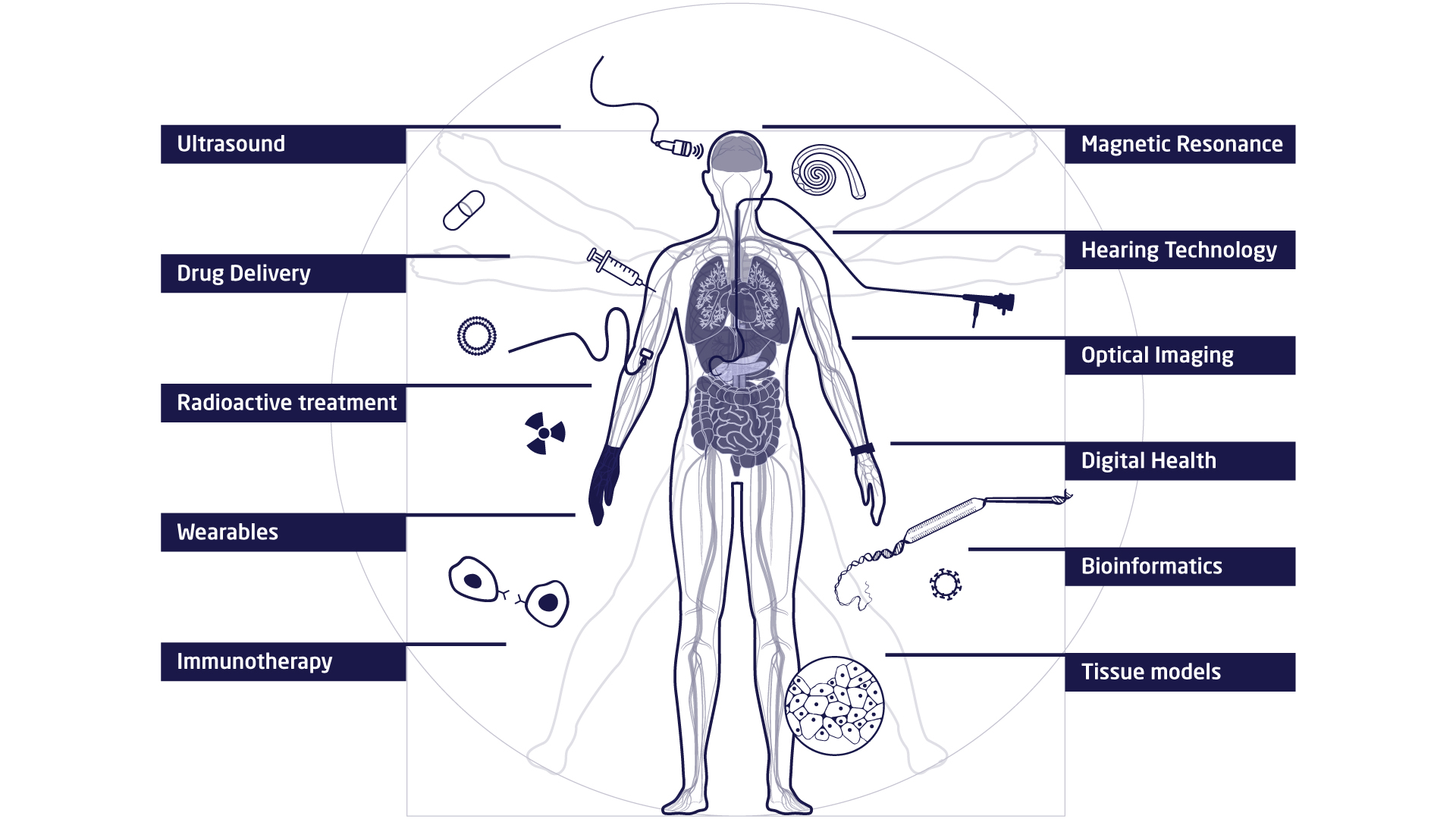
The research at DTU Health Tech is organised in research sections that each contains a number of research groups. The research sections have expertise within and across Biopharma (wet), Imaging and Diagnostics (dry) and Digital Health and Biological Modelling (data) constituting a truly interdisciplinary department. DTU Health Tech hosts a number of major research centres that include partners from industry, the health sector and academia.
Research
Research Sections
Read about the Research Sections at DTU Health Tech
Publications
READ about research topics at DTU Health Tech